About Me
I am currently a postdoctoral scholar from Department of Statistics, Stanford University, advised by Prof. Wing Hung Wong. My general research interest lies in the multi-disciplinary area where I have been committed to developing practical statistical and machine learning tools with significance in both statistical theory and applications. In particular, I have been pursuing this research agenda by exploiting the advances in generative artificial intelligence (AI) to tackle several fundamental statistical problems, such as density estimation, causal inference, and unsupervised learning with also broad applications in computational biology.
Recent News
- 2023.08 - Our Pretrained Transformer model for epigenomics is on bioRxiv.
- 2022.12 - Our Causal Inference work by deep generative modeling is on arXiv.
- 2022.10 - Our study on single cell Hi-C data clustering was accepted to Briefings in Bioinformatics.
- 2022.09 - Our study on HiChIP database was accepted to Nucleic Acids Research.
- 2022.08 - I will give a invited talk at Washington, DC for Joint Statistical Meetings (JSM) 2022.
- 2021.12 - We won the first place in NeurIPS 2021 Multimodal Single-Cell Data Integration competition two Joint Embedding tasks.
- 2021.09 - Our study on neural network boosting was accepted to NeurIPS 2021.
- 2021.03 - Our study on neural density estimation was accepted to PNAS.
More
- 2021.02 - Our study on single cell analysis with deep generative models was accepted to Nature Machine Intelligence.
- 2020.09 - Our study on molecule optimization with reinforcement learning was accepted to NeurIPS 2020.
- 2020.06 - Our study on drug sensitivity prediction with graph neural network was accepted to ECCB 2020.
- 2020.06 - Our study on gene mutation prediction using histopathological images was accepted to MICCAI 2020.
- 2019.09 - I will visit the Department of Statistics, Stanford University as a joint Ph.D. student advised by Wing Hung Wong.
- 2019.07 - I will give a invited talk at Basel, Switzerland for ISMB 2019.
- 2019.05 - I win the travel fellowship provided by International Society of Computational Biology (ISCB).
Academic Appointment

June. 2021 - Present
Department of Statistics, Stanford University, CA, USA
Postdoctoral Scholar
Education

Sep. 2019 - June. 2021
Department of Statistics, Stanford University, CA, USA
Joint Ph.D. student

Aug. 2016 - Sep. 2019
Department of Automation, Tsinghua University, Beijing, China
Ph.D. student

Aug. 2015 - Jan. 2016
Department of Computer Science, Lund University, Sweden
Exchange Student

Aug. 2012 - Jul. 2016
ShenYuan Honors College, Beihang University, Beijing, China
Bachelor of Engineering
Selected Publications
- Generative AI in Statistics -
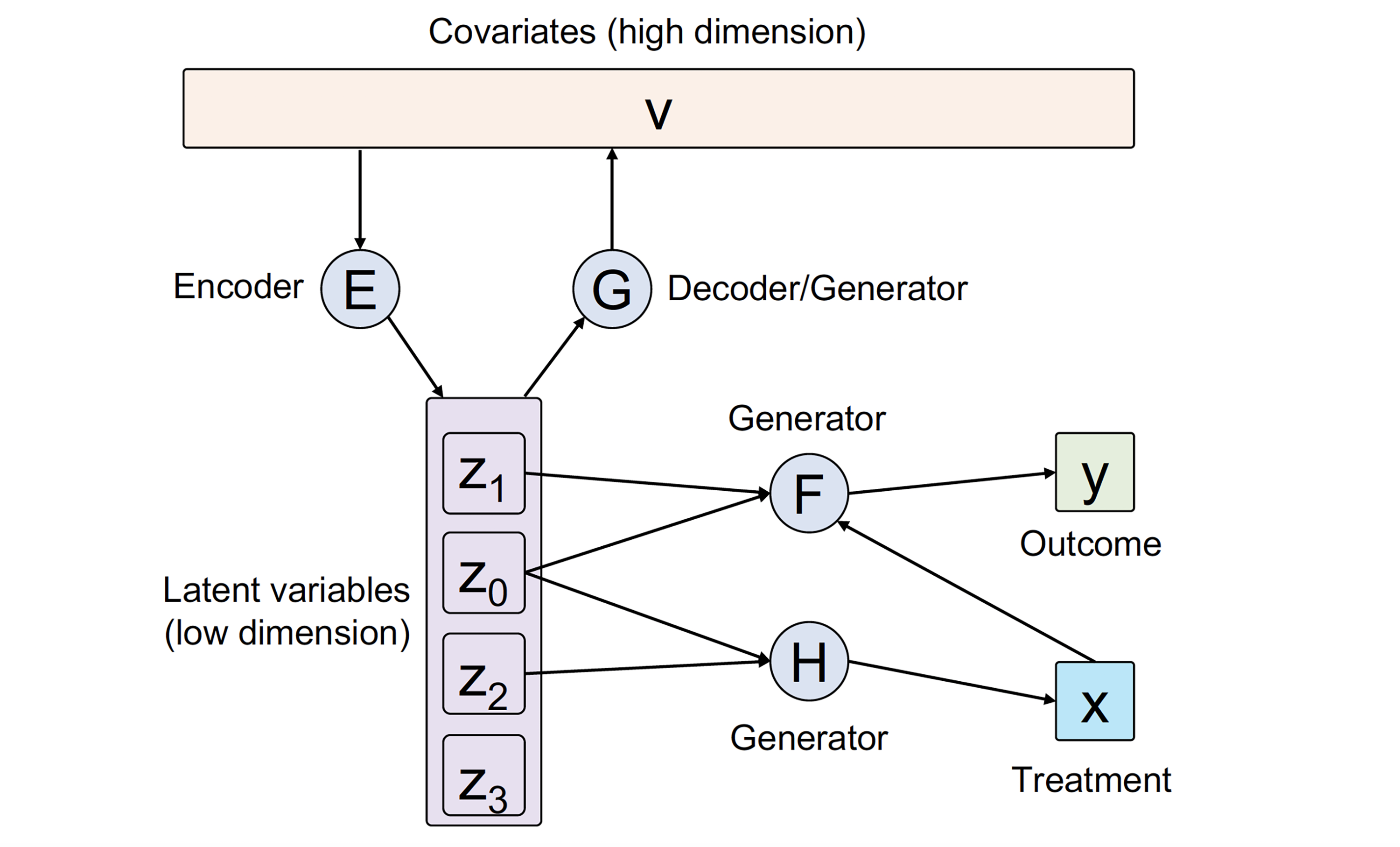 | CausalEGM: a general causal inference framework by encoding generative modeling |
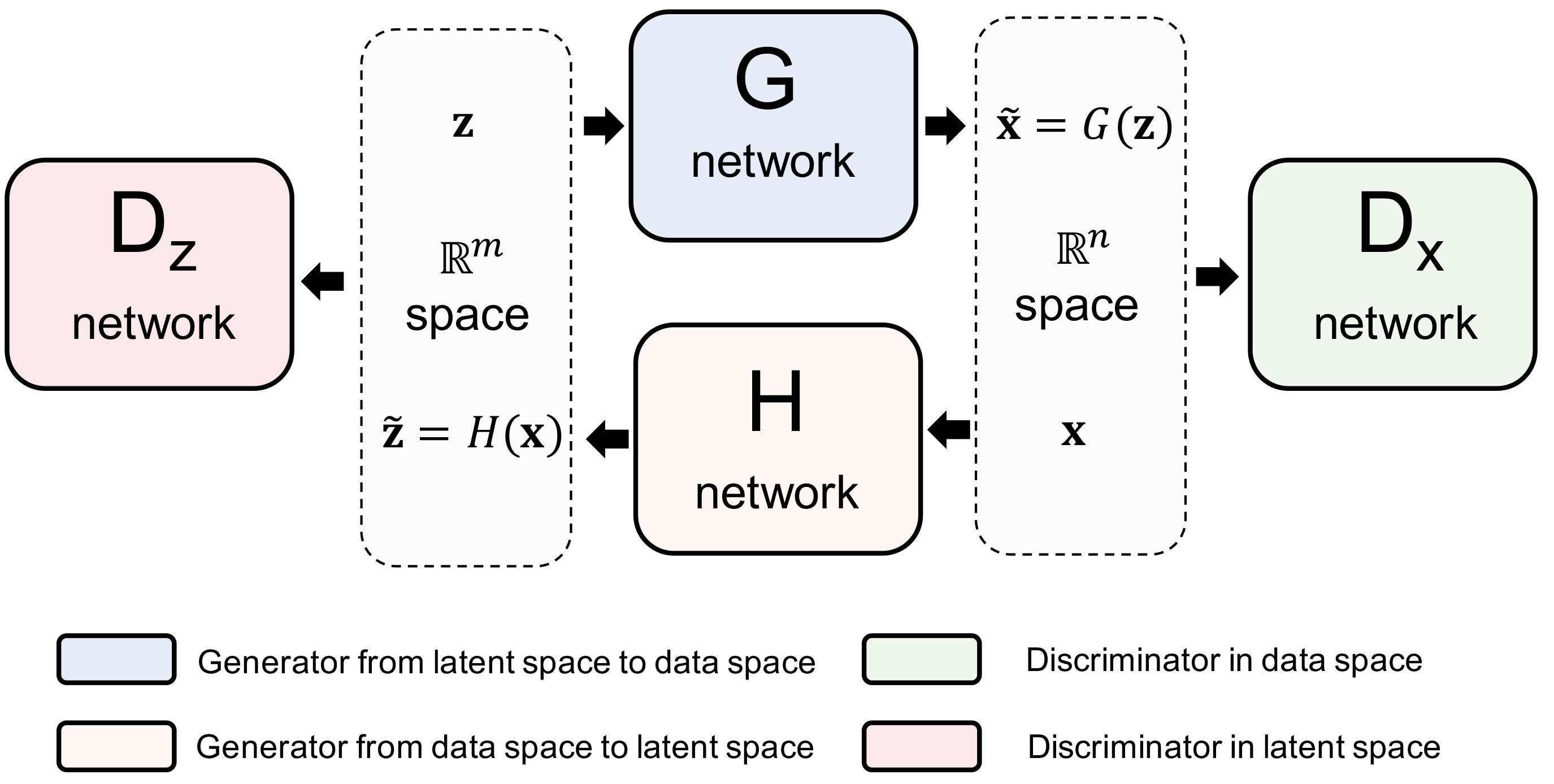 | Desity estimation with deep generative neural networks |
- Generative AI in Computational Biology -
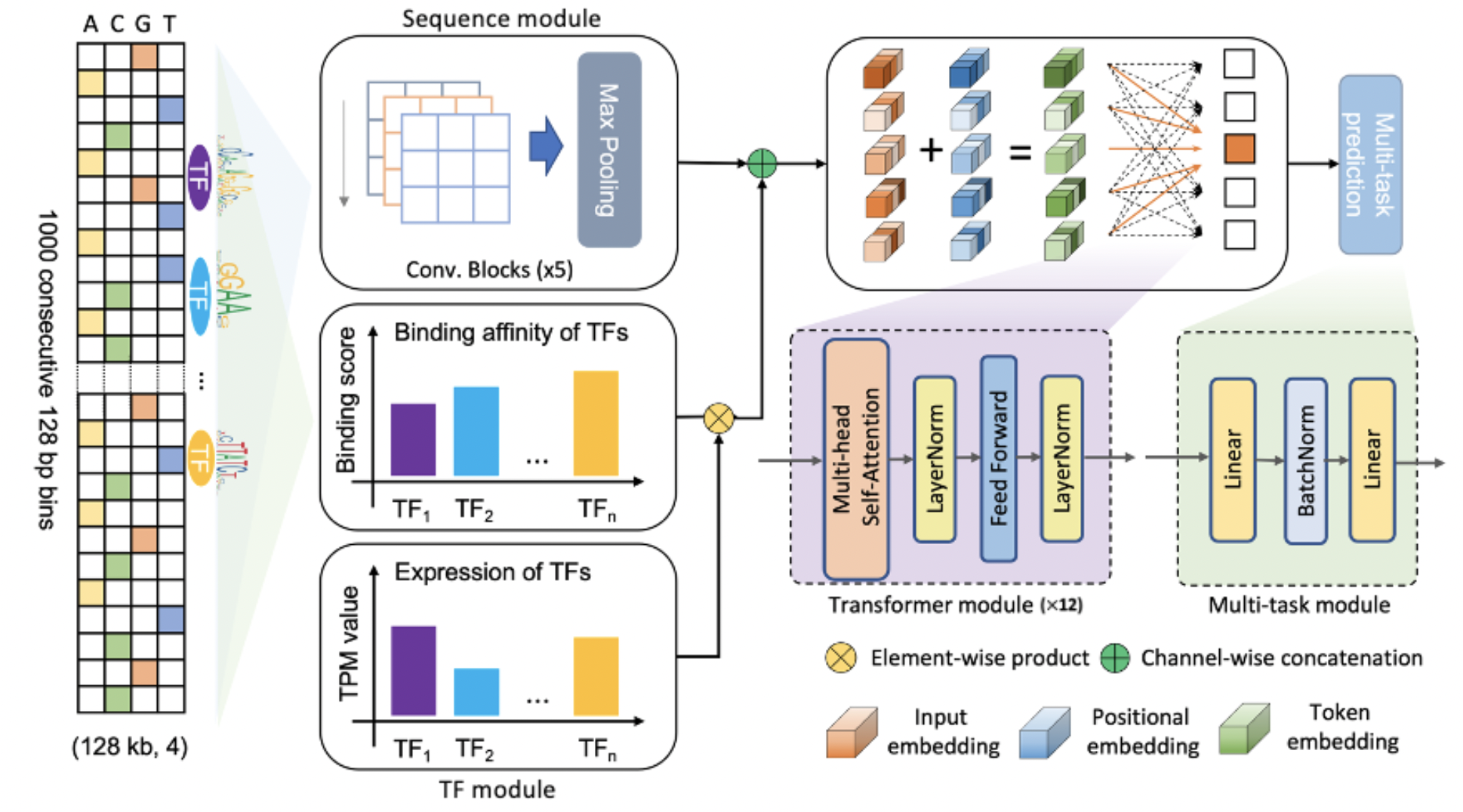 | EpiGePT: a pretrained Transformer model for epigenomics bioRxiv, 2023 [Preprint] |
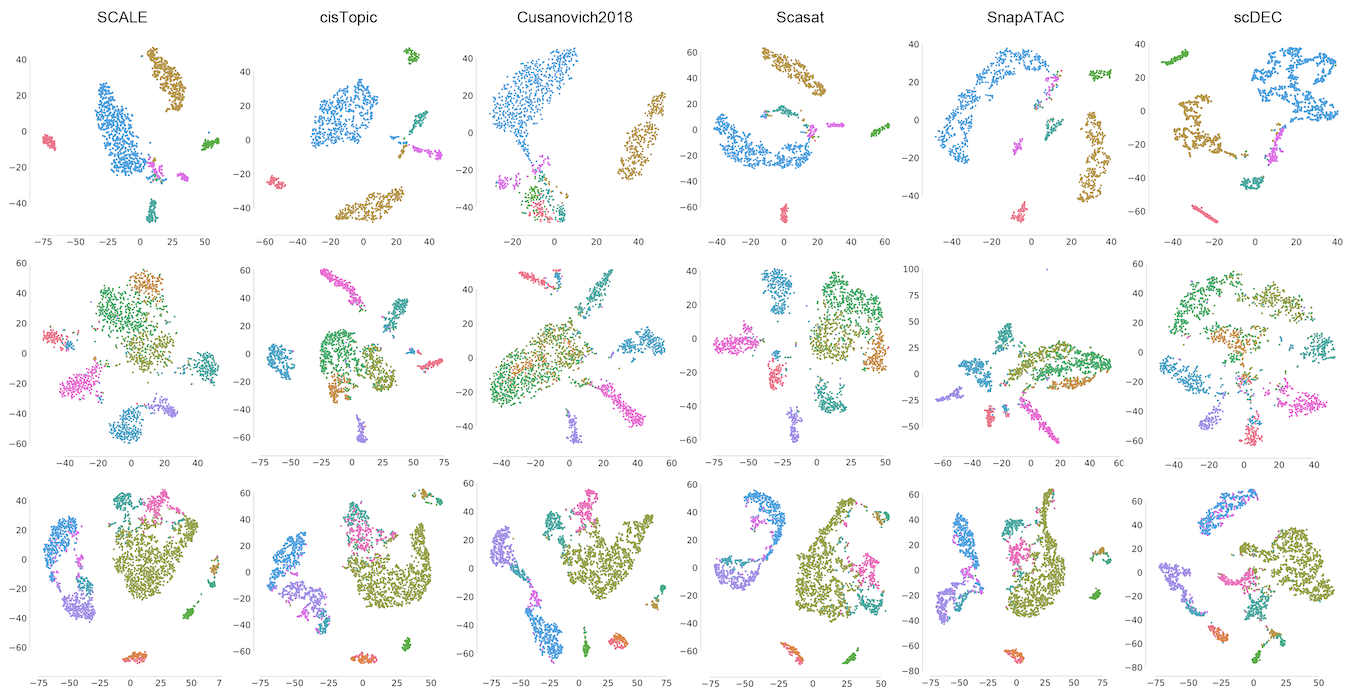 | Simultaneous deep generative modeling and clustering of single-cell genomic data |
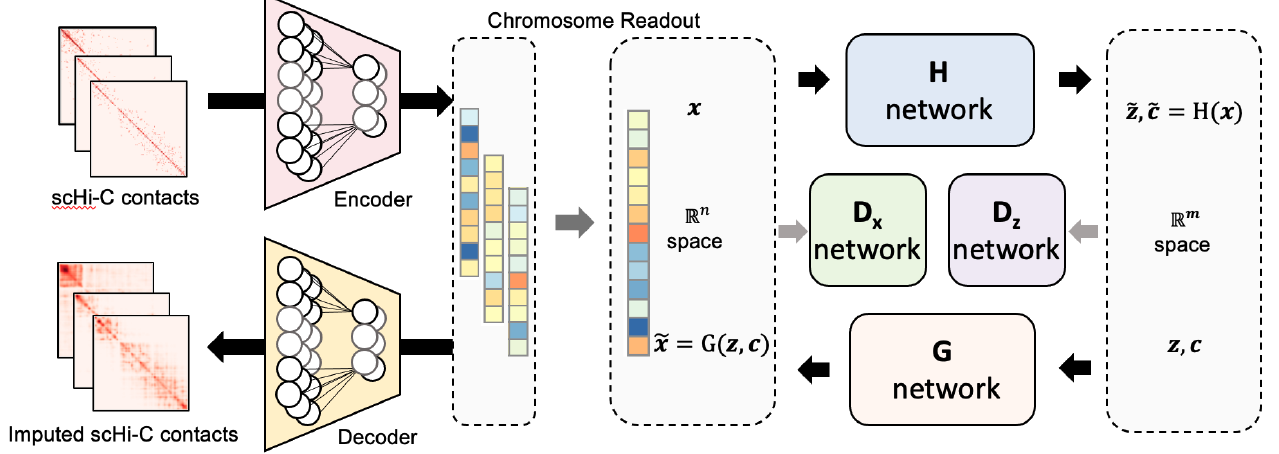 | Deep generative modeling and clustering of single cell Hi-C data Briefings in Bioinformatics, 2022 [Paper] |
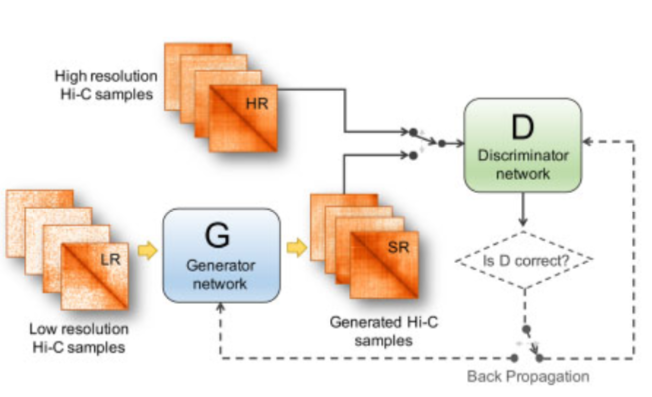 | hicGAN infers super resolution Hi-C data with generative adversarial networks |
- AI-assisted Drug Discovery -
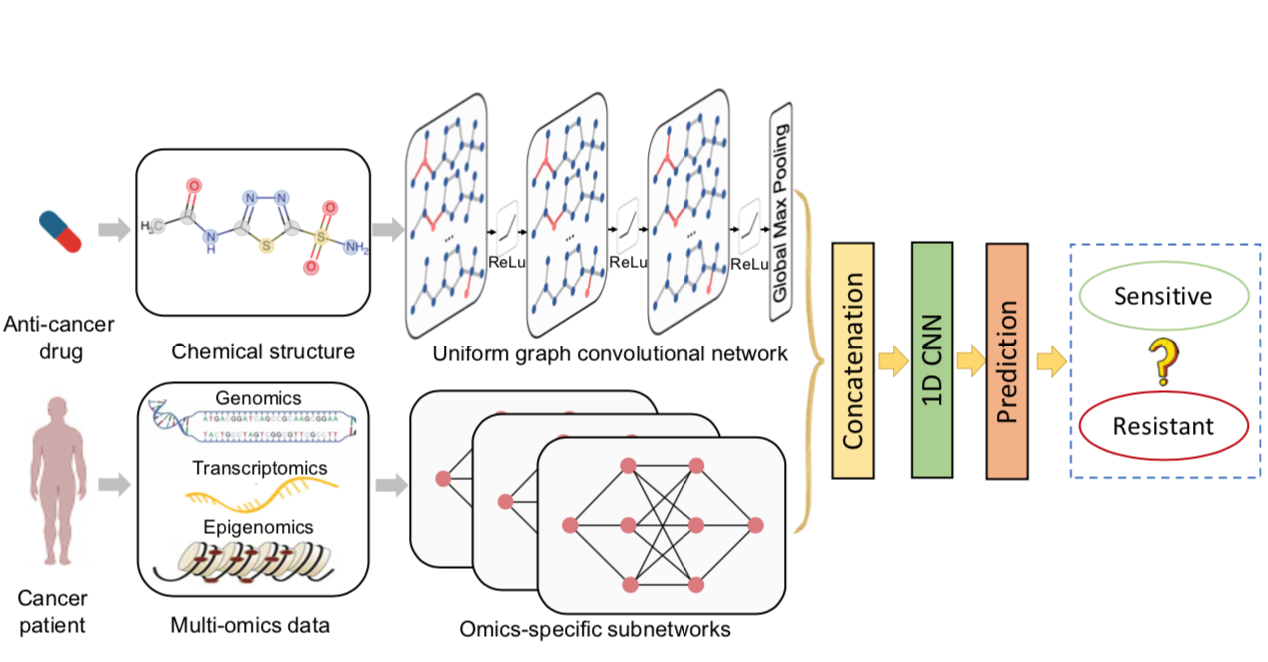 | Cancer drug response prediction via a hybrid graph convolutional network |
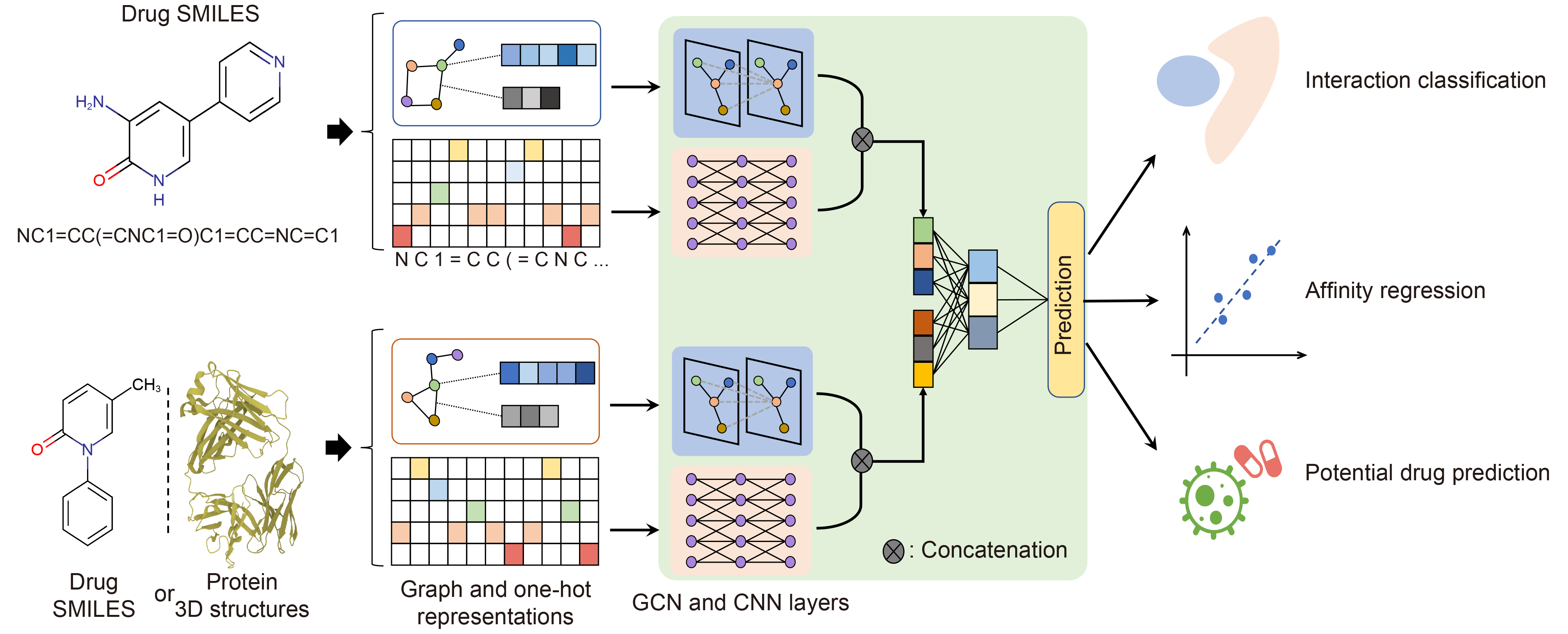 | DeepDrug: A general graph-based deep learning framework for drug-drug interactions and drug-target interactions prediction |
- Miscellaneous -
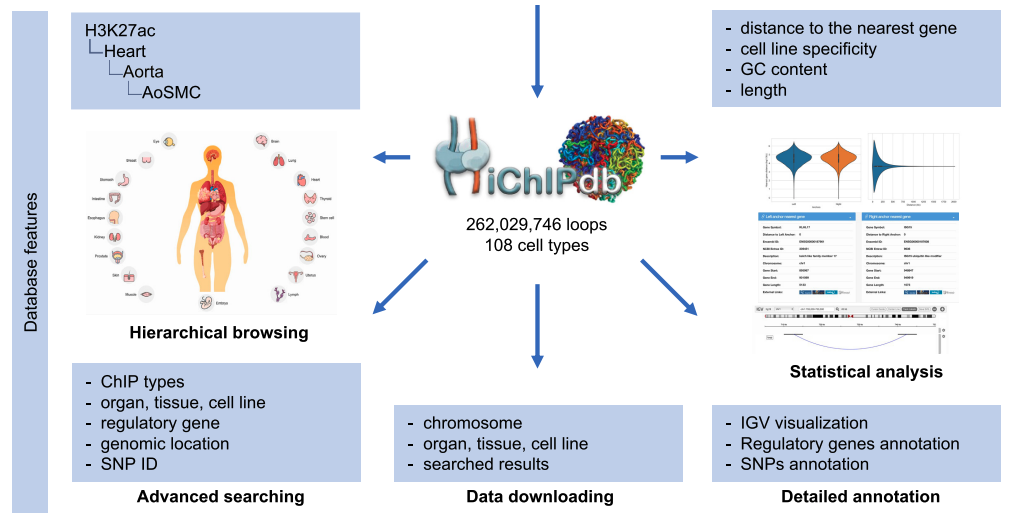 | HiChIPdb: a comprehensive database of HiChIP regulatory interactions Nucleic Acids Research, 2022 [Database Link][Paper] |
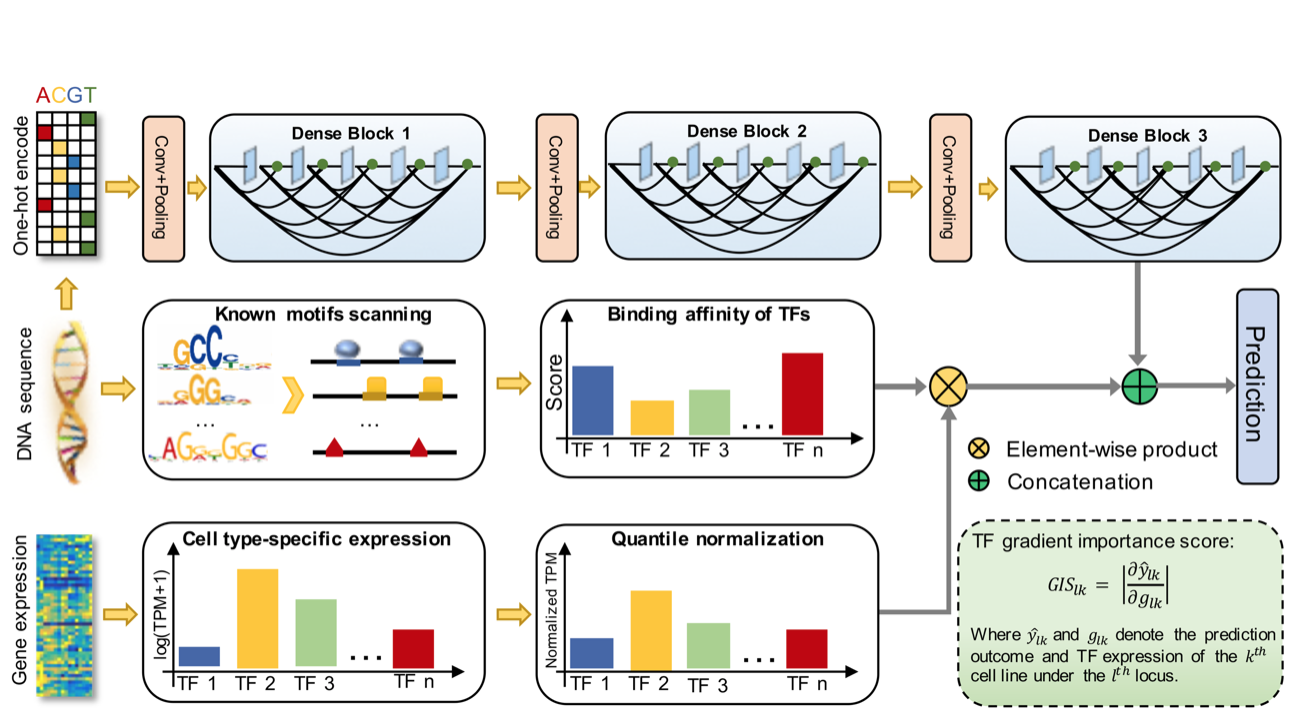 | Incorporating gene expression in genome-wide prediction of chromatin accessibility via deep learning |
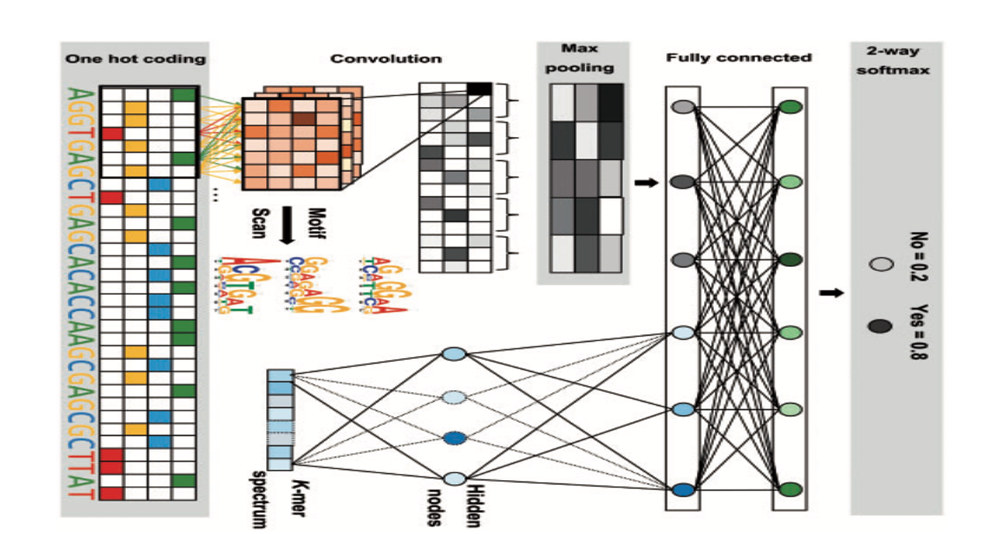 | Chromatin accessibility prediction via a hybrid deep convolutional neural network |
